My Thesis
Contents
Introduction
- Due to the abundance of drug candidates, conducting in-lab experiments to find an effective compound for a given target is a costly and time-consuming task in drug discovery.
- This thesis aims to reduce the number of drug candidates during early drug discovery by clustering the compounds.
- ChemBERTa, a Bidirectional Encoder Representation from Transformers (BERT) model, is employed to extract the descriptors for a compound.
- The compounds are clustered with respect to the learned features, and several clustering algorithms, including the k-means clustering algorithm and the Butina algorithm, are used.
- Finally, obtained clusters are evaluated by measures such as the Silhouette Score and Homogeneity Score.
- Our empirical findings show that using learned descriptors of ChemBERTa produces results that are comparable with traditional and graph-based models, as shown by metrics of cluster accuracy and computing runtime.
- Keywords: drug-target interaction, compound descriptors, representation learning, natural language processing, clustering
- Thesis Pdf
- Github Repository
Method
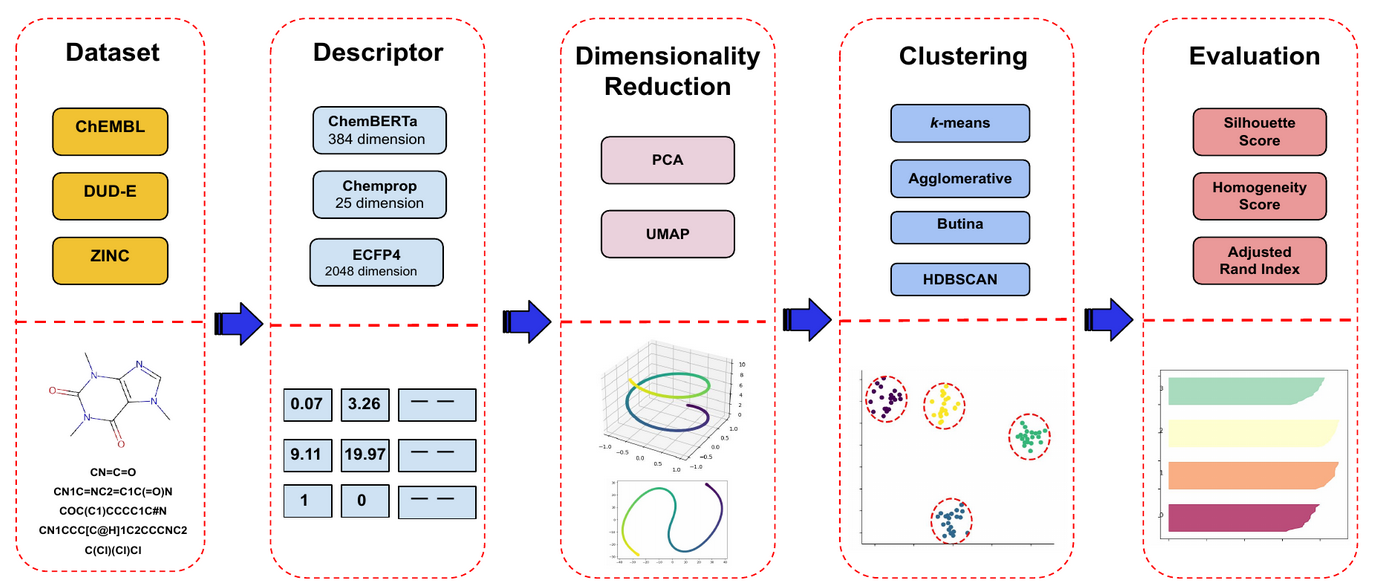
- Our method consists of 5 main stages.
- We use 3 main compound SMILES datasets, with 3 different descriptors.
- We also use dimensionality reduction techniques before clustering.
- We use 4 main clustering algorithm and evaluate their performance with 3 different metrics.
Thesis.Method